My main research projects
Mobile software development
VANTED - an advanced data and network visualization system

Under the supervision of Prof. Dr. Falk Schreiber (lead of Plant Bioinformatics group at the IPK) I created the Open Source (source code available from SourceForge) visualization system "VANTED" for the network-integrated analysis of experimental data. VANTED is the basis for most of my research and at the same time easy to use and readily available to experimentators and researchers for their visualisation and analysis work. VANTED supports Biologists and Bioinformaticians in their research. Therefore I invite you to have a look at the website http://vanted.sourceforge.net. You will find extensive documentation, a video of a example demo session which introduces the system and links to publications, which already reference VANTED as being part of their research.
Several ongoing projects have been established, which use VANTED as a
development platform. It is one of my tasks to guide and support the
developers of
these systems, mostly PhD students of our group. Therefore I provide
help on technical details,
specifications and work on the extension and adaption of the VANTED
system to enable my fellow
colleagues to work on these projects with very good results. Therefore,
the
functionality of the
system is already or will be extended by the development of Add-ons to cover various research topics of interest:
- VANTED 3D - This Add-on (developed mainly by PhD
student H.
Rohn) will enable users to work much more easily simultaneously
on large amounts of data in an intuitive way. Besides working with
*omics data and networks it is possible to integrate
3-D volumetric data, microscopic images and to visualize all of
that data in 2- and 3-D. Info: link
- SBGN-ED - the VANTED system has been extended by me and by an collaborator to support drawing of networks according to the SBGN standard, an emerging standard for drawing biological pathway data (see VANTED homepage for links to the SBGN consortium and for example drawings)
- FBA-SimVis - flux balance analysis and visualization, link
- CentiBin VANTED Add-on - a Add-on for the computation of network centralities, link
- DBE2 Add-on - development of an extended version of the DBE database, which additionally to metabolomics, genomics and proteomics data will store volume and image data, link
- MetaCrop Add-on - development of an Add-on for viewing and editing MetaCrop pathway data in a graphical (network) view, link
KGML-ED - interactive KEGG pathway navigation and visualization
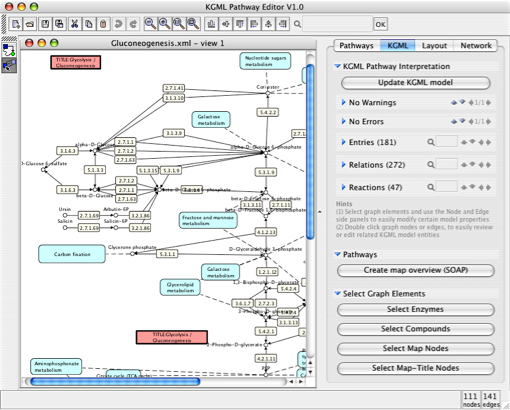
A new development, the "KGML-ED" KEGG Pathway editing system allows the visualization, exploration and editing of KEGG Pathway files. This application is a spin-off of the VANTED system, which has been modified to a large extend to support the loading and saving of KGML files and for new navigation and visualization approaches. Special requirements for the editing of KGML files required a split in the development, thus the integrated experimental data visualization is not possible with this system. The focus is on advanced pathway exploration, editing and navigation features, additionally the removal of non related user commands from the application makes it more easy to use and focused to better fit its special requirements. It is possible to easily explore several pathways in one view and integrate them into a super pathway. The resulting pathway may be saved as KGML, GML or Pajek .NET for further analysis or visualization purposes (e.g. with VANTED). Please have a look at the website http://iapg2p.sourceforge.net/kgml-ed/. You will find a short overview and tutorials which illustrate the most important features of the system.
DBE - a simple and in combination with VANTED flexible and powerful database for experiment data
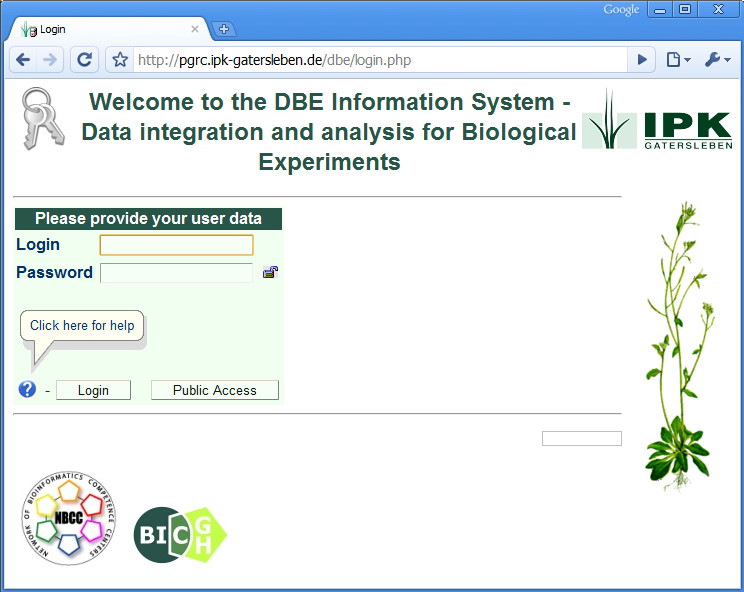
The database system "DBE", provides a web-interface for data upload and data presentation (example). It is used to store biological experiment data in a central and save place at our institute. It can also be accessed from the VANTED tool by installing a VANTED Add-on (link). Your experiment data can be easily shared and combined with different datasets using the VANTED and DBE systems.
3-D reconstruction - automatic 3-D model generation by analyzing the structure of object rotation pictures
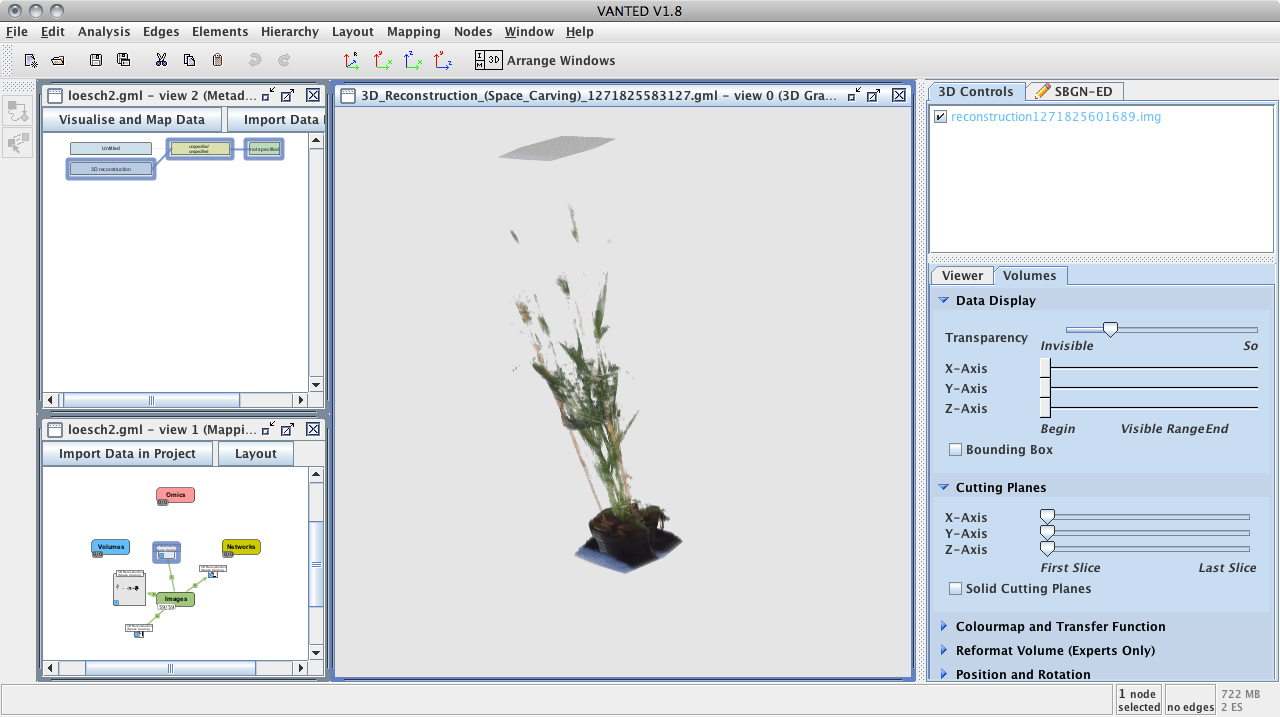
A newly developed approach for the automatic 3-D model generation has been developmented and can be used to analyze and visualize the development of plants using space-carving technology (integrated in IAP).
IAP - the Integrated Analysis Platform
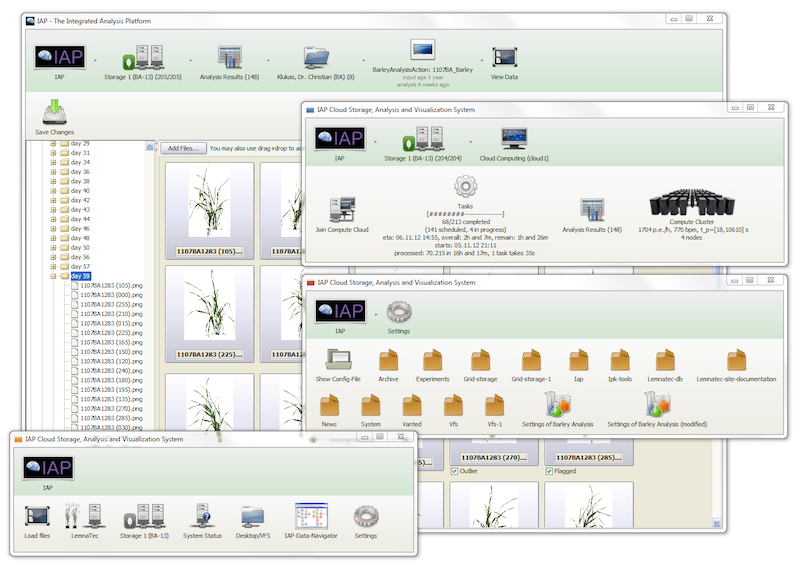
This information system will provides access to large-scale high-throughput image data, generated by automatic plant phenotyping systems. Advanced, still easy to use image analysis commands enable researchers to store, manage, retrieve, analyse and present plant phenotypic images, as well as related metabolomics, proteomics or genomics information. Therefore, this information system could become an important hub for systems biology oriented research.
IAP development has been completed, is available for download and has been published. At its core the system is based on VANTED.

















